Differences between pangenome and panproteome
PanTools offers functionalities to build and analyze a pangenome or panproteome.
A pangenome is constructed from genome and annotation files. First, genome sequences are k-merized and compressed into a De Bruijn graph. Genes and other annotation features from annotation files are integrated into the pangenome as ‘gene’, ‘mRNA’ and ‘CDS’ nodes. Gene start and stop positions are annotated in the graph as relationships and connect the annotation layer to the nucleotide layer. The protein sequences can be clustered into homology groups and connect homologous proteins from different genomes.
A panproteome is built from protein sequences only, ignoring the underlying genome structure. Again, the protein sequences are clustered into homology groups which serve as main input for many functionalities.
In addition to the single layer in panproteomes and three layers in pangenomes, a functional layer can be included in both databases. This layer consists of multiple functional annotation databases (e.g. GO, PFAM) and connects proteins with a shared function.
Since there is only a protein layer and functional layer present in panproteomes, not all functions can be utilized. See the table below for which functions can be used for pangenomes and panproteomes.
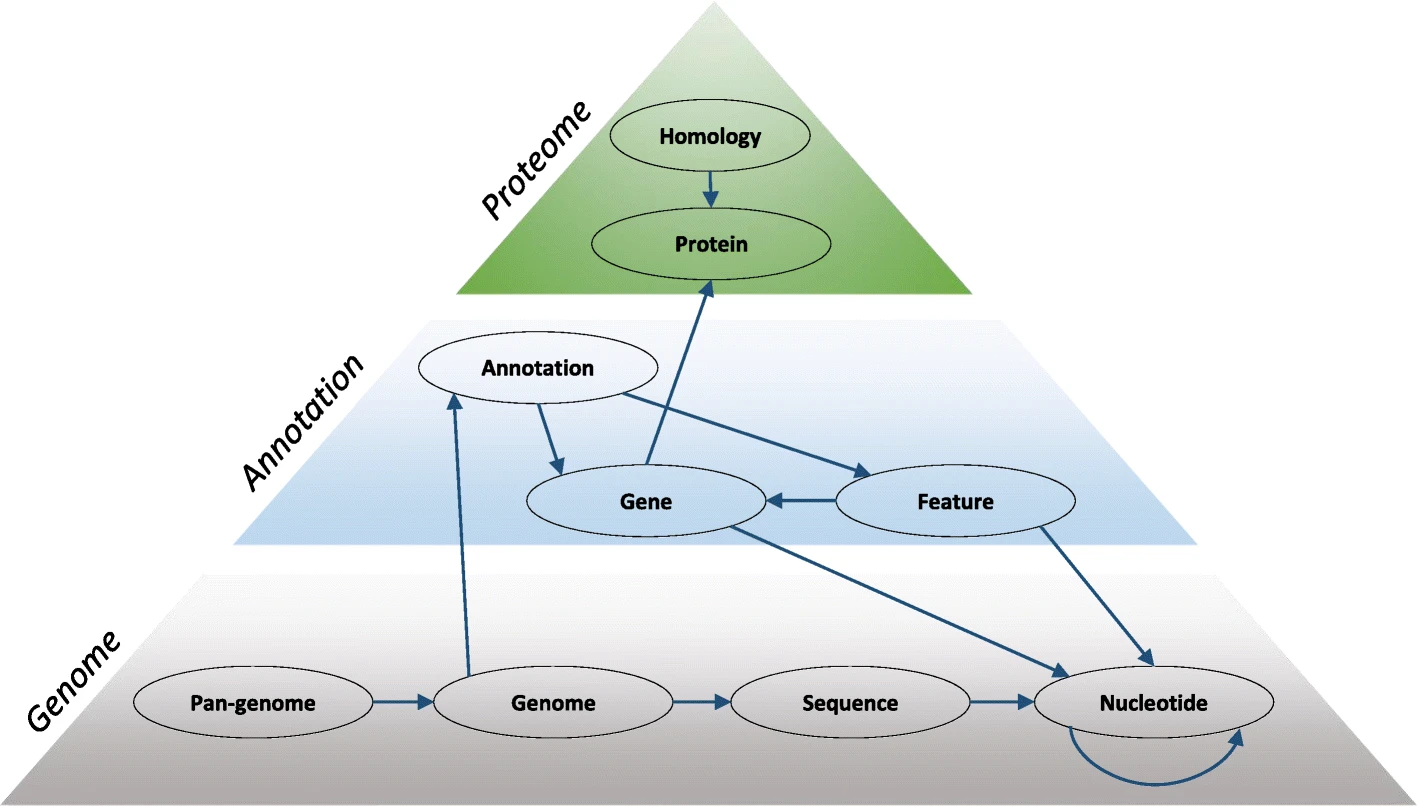
Fig. 1 Schematic of genome, annotation, and protein layer of a pangenome database. Figure taken from Efficient inference of homologs in large eukaryotic pan-proteomes
Available functions
Pangenome construction and annotation
Function |
Pangenome |
Panproteome |
|---|---|---|
build_pangenome |
YES |
NO |
add_genomes |
YES |
NO |
build_panproteome |
NO |
YES |
add_annotations |
YES |
NO |
remove_annotations |
YES |
NO |
add_functions |
YES |
YES |
remove_functions |
YES |
YES |
add_antismash |
YES |
NO |
function_overview |
YES |
YES |
add_phenotype |
YES |
YES |
remove_phenotype |
YES |
YES |
add_variants |
YES |
NO |
remove_variants |
YES |
NO |
add_pavs |
YES |
YES |
remove_pavs |
YES |
YES |
variation_overview |
YES |
YES |
remove_nodes |
YES |
YES |
add_phasing |
YES |
NO |
add_repeats |
YES |
NO |
repeat_overview |
YES |
NO |
group |
YES |
YES |
busco_protein |
YES |
YES |
optimal_grouping |
YES |
YES |
change_grouping |
YES |
YES |
deactivate_grouping |
YES |
YES |
remove grouping |
YES |
YES |
calculate_synteny |
YES |
NO |
add_synteny |
YES |
NO |
synteny_overview |
YES |
NO |
Pangenome analysis
Function |
Pangenome |
Panproteome |
|---|---|---|
blast |
YES |
NO |
gene_classification |
YES |
YES |
core_unique_thresholds |
YES |
YES |
pangenome_structure |
YES |
YES |
kmer_classification |
YES |
NO |
functional_classification |
YES |
YES |
locate_genes |
YES |
NO |
find_genes_by_name |
YES |
NO |
find_genes_by_annotation |
YES |
NO |
find_genes_in_region |
YES |
NO |
show_go |
YES |
YES |
compare_go |
YES |
YES |
group_info |
YES |
YES |
retrieve_regions |
YES |
NO |
retrieve_features |
YES |
NO |
gene_retention |
YES |
NO |
go_enrichment |
YES |
YES |
order_matrix |
YES |
YES |
rename_matrix |
YES |
YES |
metrics |
YES |
YES |
msa |
YES |
YES |
core_phylogeny |
YES |
YES |
consensus_tree |
YES |
YES |
ani |
YES |
NO |
mlsa_find_genes |
YES |
NO |
mlsa_concatenate |
YES |
NO |
mlsa |
YES |
NO |
edit_phylogeny |
YES |
YES |
root_phylogeny |
YES |
YES |
create_tree_template |
YES |
YES |
map |
YES |
NO |
sequence_visualization |
YES |
NO |
calculate_dn_ds |
YES |
NO |