Annotate the pangenome graph
Structural annotations
Add annotations
Construct or expand the annotation layer of an existing pangenome. The layer consists of genomic features like genes, mRNAs, proteins, tRNAs etc. PanTools is only able to read General Feature Format (GFF) files.
Multiple annotations can be assigned to a single genome; however, only
one annotation a time can be included in an analysis. The most recently
included annotation of a genome is included as default, unless a
different annotation is specified via --annotations-file. This annotation
file contains only annotation identifiers, each on a separate line. The most
recent annotation is used for genomes where no annotation number is specified
in the file. Below is an example where the third annotation of genome 1 is
selected and the second annotation of genome 2 and 3.
1_3
2_2
3_2
- Parameters
<databaseDirectory>
Path to the database root directory.
<annotationsFile>
A text file with on each line a genome number and the full path to the corresponding annotation file, separated by a space.
- Options
--connectConnect the annotated genomic features to nucleotide nodes in the DBG.
--ignore-invalid-featuresIgnore GFF3 features that do not match the fasta.
--assume-one-mrna-per-cdsOnly relevant for features in GFF files that lack an mRNA between CDS and gene. By default, PanTools will assume that all CDS features belong to the same mRNA. If this option is set, PanTools will assume that each CDS feature belongs to a separate mRNA. For most GFF files this option should not be set.
- Example commands
$ pantools add_annotations tomato_DB annotations.txt $ pantools add_annotations --connect tomato_DB annotations.txt
- Output
The annotated features are incorporated in the graph. Output files are written to the database directory.
annotation_overview.txt, a summary of the GFF files incorporated in the pangenome.
annotation.log, a list of misannotated feature identifiers.
- Example input file
Each line of the file starts with the genome number followed by the full path to the annotation file. The genome numbers match the line number of the file that you used to construct the pangenome.
1 /always/genome1.gff 2 /use_the/genome2.gff 3 /full_path/genome3.gff
- GFF3 file format
The GFF format consists of one line per feature, each containing 9 columns of data, plus optional track definition lines, that must be tab separated. Please use the proper hierarchy for the feature: gene -> mRNA -> CDS. Where gene is the parent of mRNA and mRNA is the parent of the CDS feature. The following example from Saccharomyces cerevisiae YJM320 (GCA_000975885) displays a correctly formatted gene entry:
CP004621.1 Genbank gene 44836 45753 . - . ID=gene99;Name=RPL23A;end_range=45753,.;gbkey=Gene;gene=RPL23A;gene_biotype=protein_coding;locus_tag=H754_YJM320B00023;partial=true;start_range=.,44836 CP004621.1 Genbank mRNA 44836 45753 . - . ID=rna99;Parent=gene99;gbkey=mRNA;gene=RPL23A;product=Rpl23ap CP004621.1 Genbank exon 45712 45753 . - . ID=id112;Parent=rna99;gbkey=mRNA;gene=RPL23A;product=Rpl23ap CP004621.1 Genbank exon 44836 45207 . - . ID=id113;Parent=rna99;gbkey=mRNA;gene=RPL23A;product=Rpl23ap CP004621.1 Genbank CDS 45712 45753 . - 0 ID=cds92;Parent=rna99;Dbxref=SGD:S000000183,NCBI_GP:AJQ01854.1;Name=AJQ01854.1;Note=corresponds to s288c YBL087C;gbkey=CDS;gene=RPL23A;product=Rpl23ap;protein_id=AJQ01854.1 CP004621.1 Genbank CDS 44836 45207 . - 0 ID=cds92;Parent=rna99;Dbxref=SGD:S000000183,NCBI_GP:AJQ01854.1;Name=AJQ01854.1;Note=corresponds to s288c YBL087C;gbkey=CDS;gene=RPL23A;product=Rpl23ap;protein_id=AJQ01854.1
--annotations-file is included specifying which annotations to use.
This annotation file contains only annotation identifiers, each on a
separate line. The most recent annotation is used for genomes where no
annotation number is specified in the file. Below is an example where
the third annotation of genome 1 is selected and the second annotation
of genome 2 and 3.1_3
2_2
3_2
Remove annotations
Remove all the genomic features that belong to annotations, such as gene, mRNA, exon, tRNA, and feature nodes. Functional annotation nodes are not removed with this function but can be removed with remove_functions. Removing annotations can be done in two ways:
Selecting genomes with
--includeor--exclude, for which all annotation features will be removed.Remove specific annotations by providing a text file with identifiers via the
--annotations-fileargument.
- Parameters
<databaseDirectory>
Path to the database root directory.
- Options
Requires one of
--include|--exclude|--annotations-file.--include/-iA selection of genomes for which all annotations will be removed.
--exclude/-eA selection of genomes excluded from the removal of annotations.
--annotations-file/-AA text file with the identifiers of annotations to be removed, each on a separate line.
- Example annotations file
The annotations file should contain identifiers for annotations on each line (genome number, annotation number). The following example will remove the first annotations of genome 1, 2 and 3 and the second annotation of genome 1.
1_1 1_2 2_1 3_1
- Example commands
$ pantools remove_annotations --exclude=3,4,5 $ pantools remove_annotations -A annotations.txt
Functional annotations
PanTools is able to incorporate functional annotations into the pangenome by reading output from various functional annotation tools.
Add functions
This function can integrate different functional annotations from a variety of annotation files. Currently available functional annotations: Gene Ontology, Pfam, InterPro, TIGRFAM, Phobius, SignalP and COG. The first time this function is executed, the Pfam, TIRGRAM, GO, and InterPro databases are integrated into the pangenome. Phobius, SignalP and COG annotations do not have separate nodes and are directly annotated on ‘mRNA’ nodes in the pangenome.
Gene names (or identifiers) from the input file are used to identify
gene nodes in the pangenome. Only genes with an exactly matching
name/identifier can be connected to functional annotation nodes! Use the
same FASTA and GFF3 files that were used to construct the pangenome database.
(It is best to use the protein fasta files in the proteins directory of the
database.)
-F option to prevent
unnecessary downloads from the internet, preferably to a location easily
accessible.Database type |
Version |
|---|---|
GO |
2021-12-15 |
Pfam |
35.0 |
TIGRFAM |
15.0 |
InterPro |
87.0 |
File |
Database type |
Download link |
|---|---|---|
go.basic.obo |
GO |
|
gene_ontology.txt |
Pfam |
ftp://ftp.ebi.ac.uk/pub/databases/Pfam/releases//Pfam35.0/database_files/gene_ontology.txt.gz |
Pfam-A.clans.tsv |
Pfam |
ftp://ftp.ebi.ac.uk/pub/databases/Pfam/releases//Pfam35.0/Pfam-A.clans.tsv.gz |
interpro.xml |
InterPro |
https://ftp.ebi.ac.uk/pub/databases/interpro/current_release/interpro.xml.gz |
TIGRFAMS_GO_LINK |
TIGRFAM |
https://ftp.ncbi.nlm.nih.gov/hmm/TIGRFAMs/release_15.0/TIGRFAMS_GO_LINK |
TIGRFAMS_ROLE_LINK |
TIGRFAM |
https://ftp.ncbi.nlm.nih.gov/hmm/TIGRFAMs/release_15.0/TIGRFAMS_ROLE_LINK |
TIGR_ROLE_NAMES |
TIGRFAM |
https://ftp.ncbi.nlm.nih.gov/hmm/TIGRFAMs/release_15.0/TIGR_ROLE_NAMES |
TIGR00001.INFO to TIGR04571.INFO |
TIGRFAM |
https://ftp.ncbi.nlm.nih.gov/hmm/TIGRFAMs/release_15.0/TIGRFAMs_15.0_INFO.tar.gz |
- Parameters
<databaseDirectory>
Path to the database root directory.
<functionsFile>
A text file with on each line a genome number and the full path to the corresponding annotation file, separated by a space.
- Options
--annotations-file/-AA text file with the identifiers of annotations to be included, each on a separate line. The most recent annotation is selected for genomes without an identifier.
--functional-databases-directory/-FPath to the directory containing the functional databases. If the databases are not present, they are downloaded automatically. (Default location is “functional_databases” in the database directory.)
- Example commands
$ pantools add_functions -F ~/function_databases tomato_DB f_annotations.txt $ pantools add_functions -F ~/function_databases -A annotations.txt tomato_DB f_annotations.txt
- Output
Functional annotations are incorporated in the graph. A log file is written to the log directory.
add_functional_annotations.log, a log file with the the number of added functions per type and the identifiers of functions that could not be included.
- Example function files
The <functionsFile> requires to be formatted like an annotation input file. Each line of the file starts with the genome number followed by the full path to an annotation file.
File type
Recognized by pattern in file name
InterProScan
interpro & .gff
eggNOG-mapper
eggnog
Phobius
phobius
SignalP
signalp
Custom file
custom
1 /mnt/scratch/interpro_results_genome_1.gff 1 /mnt/scratch/custom_annotation_1.txt 1 /mnt/scratch/phobius_1.txt 2 /mnt/scratch/signalp.txt 2 /mnt/scratch/eggnog_genome_2.annotations 2 /mnt/scratch/transmembrane_annotations.txt phobius 3 /mnt/scratch/ipro_results_genome_3.annot custom
- Annotation file types
PanTools can recognize functional annotations in different output formats.
Phobius and SignalP are not standard analyses of the InterProScan pipeline and require some additional steps during the InterProScan installation. Please take a look at our InterProScan install instruction to verify if the tools are part of the prediction pipeline. Phobius 1.01
Function type
Allowed annotation file
GO
InterProscan .gff & custom annotation file
Pfam
InterProscan .gff & custom annotation file
InterPro
InterProscan .gff & custom annotation file
TIGRFAM
InterProscan .gff & custom annotation file
Phobius
InterProscan .gff & Phobius 1.01 output
SignalP
InterProscan .gff, signalP 4.1 output, signalP 5.0 output
COG
eggNOG-mapper
InterProScan gff file:
##gff-version 3 ##interproscan-version 5.52-86.0 AT4G21230.1 ProSiteProfiles protein_match 333 620 39.000664 + . date=06-10-2021;Target=mRNA.AT4G21230.1 333 620;Ontology_term="GO:0004672","GO:0005524","GO:0006468";ID=match$42_333_620;signature_desc=Protein kinase domain profile.;Name=PS50011;status=T;Dbxref="InterPro:IPR000719" AT3G08980.5 TIGRFAM protein_match 25 101 3.7E-14 + . date=06-10-2021;Target=mRNA.AT3G08980.5 25 101;Ontology_term="GO:0006508","GO:0008236","GO:0016020";ID=match$66_25_101;signature_desc=sigpep_I_bact: signal peptidase I;Name=TIGR02227;status=T;Dbxref="InterPro:IPR000223" AT2G17780.2 Phobius protein_match 338 354 . + . date=06-10-2021;Target=AT2G17780.2 338 354;ID=match$141_338_354;signature_desc=Region of a membrane-bound protein predicted to be embedded in the membrane.;Name=TRANSMEMBRANE;status=T AT2G17780.2 Phobius protein_match 1 337 . + . date=06-10-2021;Target=AT2G17780.2 1 337;ID=match$142_1_337;signature_desc=Region of a membrane-bound protein predicted to be outside the membrane, in the extracellular region.;Name=NON_CYTOPLASMIC_DOMAIN;status=T AT3G11780.2 SignalP_EUK protein_match 1 24 . + . date=06-10-2021;Target=mRNA.AT3G11780.2 1 24;ID=match$230_1_24;Name=SignalP-noTM;status=T AT1G04300.2 CDD protein_match 40 114 1.54717E-13 + . date=06-10-2021;Target=mRNA.AT1G04300.2 40 114;Ontology_term="GO:0005515";ID=match$212_40_114;signature_desc=MATH;Name=cd00121;status=T;Dbxref="InterPro:IPR002083"
eggNOG-mapper (tab separated) file:
#query_name seed_eggNOG_ortholog seed_ortholog_evalue seed_ortholog_score best_tax_level Preferred_name GOs EC KEGG_ko KEGG_Pathway KEGG_Module KEGG_Reaction KEGG_rclass BRITE KEGG_TC CAZy BiGG_Reaction taxonomic scope eggNOG OGs best eggNOG OG COG Functional cat. eggNOG free text desc. ATKYO-2G54530.1 3702.AT2G35130.2 1.9e-179 636.0 Brassicales GO:0003674,GO:0003676,GO:0003723,GO:0003824,GO:0004518,GO:0004519,GO:0005488,GO:0005575,GO:0005622,GO:0005623,GO:0006139,GO:0006725,GO:0006807,GO:0008150,GO:0008152,GO:0009451,GO:0009987,GO:0016070,GO:0016787,GO:0016788,GO:0034641,GO:0043170,GO:0043226,GO:0043227,GO:0043229,GO:0043231,GO:0043412,GO:0044237,GO:0044238,GO:0044424,GO:0044464,GO:0046483,GO:0071704,GO:0090304,GO:0090305,GO:0097159,GO:1901360,GO:1901363 Viridiplantae 37R67@33090,3GAUT@35493,3HNDD@3699,KOG4197@1,KOG4197@2759 NA|NA|NA E Pentacotripeptide-repeat region of PRORP ATKYO-UG22500.1 3712.Bo02269s010.1 7.5e-35 153.7 Brassicales Viridiplantae 29I9W@1,2RRH4@2759,383W6@33090,3GWQZ@35493,3I1A9@3699 NA|NA|NA ATKYO-1G60060.1 3702.AT1G48090.1 0.0 6241.0 Brassicales ko:K19525 ko00000 Viridiplantae 37IJB@33090,3GAN0@35493,3HQ90@3699,COG5043@1,KOG1809@2759 NA|NA|NA U Vacuolar protein sorting-associated protein ATKYO-3G74720.1 3702.AT3G52120.1 7.2e-245 852.8 Brassicales ko:K13096 ko00000,ko03041 Viridiplantae 37QYY@33090,3G9VU@35493,3HRDK@3699,KOG0965@1,KOG0965@2759 NA|NA|NA L SWAP (Suppressor-of-White-APricot) surp domain-containing protein D111 G-patch domain-containing protein ATKYO-4G41660.1 3702.AT4G16340.1 0.0 3392.1 Brassicales GO:0003674,GO:0005085,GO:0005088,GO:0005089,GO:0005488,GO:0005515,GO:0005575,GO:0005622,GO:0005623,GO:0005634,GO:0005737,GO:0005783,GO:0005829,GO:0005886,GO:0006810,GO:0008064,GO:0008150,GO:0008360,GO:0009605,GO:0009606,GO:0009628,GO:0009629,GO:0009630,GO:0009958,GO:0009966,GO:0009987,GO:0010646,GO:0010928,GO:0012505,GO:0016020,GO:0016043,GO:0016192,GO:0017016,GO:0017048,GO:0019898,GO:0019899,GO:0022603,GO:0022604,GO:0023051,GO:0030832,GO:0031267,GO:0032535,GO:0032956,GO:0032970,GO:0033043,GO:0043226,GO:0043227,GO:0043229,GO:0043231,GO:0044422,GO:0044424,GO:0044425,GO:0044432,GO:0044444,GO:0044446,GO:0044464,GO:0048583,GO:0050789,GO:0050793,GO:0050794,GO:0050896,GO:0051020,GO:0051128,GO:0051179,GO:0051234,GO:0051493,GO:0065007,GO:0065008,GO:0065009,GO:0070971,GO:0071840,GO:0071944,GO:0090066,GO:0098772,GO:0110053,GO:1902903 ko:K21852 ko00000,ko04131 Viridiplantae 37QIM@33090,3G8RK@35493,3HSFN@3699,KOG1997@1,KOG1997@2759 NA|NA|NA T Belongs to the DOCK family
A custom input file must consist of two tab or comma separated columns. The first column should contain a gene/mRNA id, the second an identifier from one of four functional annotation databases: GO, Pfam, InterPro or TIGRFAM.
AT5G23090.4,GO:0046982 AT5G23090.4,IPR009072 AT1G27540.2,PF03478 AT2G18450.1,TIGR01816
Phobius 1.01 ‘short’ (tab separated) functions file:
SEQENCE ID TM SP PREDICTION mRNA-YPR204W 0 0 o mRNA-ndhB-2_1 6 Y n5-16c21/22o37-57i64-83o89-113i134-156o168-189i223-246o
Phobius 1.01 ‘long’ (tab separated) functions file:
ID mRNA-YPR204W FT DOMAIN 1 1032 NON CYTOPLASMIC. // ID mRNA-ndhB-2_1 FT SIGNAL 1 21 FT DOMAIN 1 4 N-REGION. FT DOMAIN 5 16 H-REGION. FT DOMAIN 17 21 C-REGION. FT DOMAIN 22 36 NON CYTOPLASMIC. FT TRANSMEM 37 57 FT DOMAIN 58 63 CYTOPLASMIC. FT TRANSMEM 64 83 FT DOMAIN 84 88 NON CYTOPLASMIC. FT TRANSMEM 89 113 FT DOMAIN 114 133 CYTOPLASMIC. FT TRANSMEM 134 156 FT DOMAIN 157 167 NON CYTOPLASMIC. FT TRANSMEM 168 189 FT DOMAIN 190 222 CYTOPLASMIC. FT TRANSMEM 223 246 FT DOMAIN 247 253 NON CYTOPLASMIC. //
SignalP 4.1 ‘short’ (tab separated) functions file:
# name Cmax pos Ymax pos Smax pos Smean D ? Dmaxcut Networks-used mRNA-rpl2-3 0.148 20 0.136 20 0.146 3 0.126 0.131 N 0.450 SignalP-noTM mRNA-cox2 0.107 25 0.132 12 0.270 4 0.162 0.148 N 0.450 SignalP-noTM mRNA-cox2_1 0.850 17 0.776 17 0.785 2 0.717 0.753 Y 0.500 SignalP-TM
SignalP 5.0 ‘short’ (tab separated) functions file:
# SignalP-5.0 Organism: Eukarya Timestamp: 20211122233246 # ID Prediction SP(Sec/SPI) OTHER CS Position AT3G26880.1 SP(Sec/SPI) 0.998803 0.001197 CS pos: 21-22. VYG-KK. Pr: 0.9807 mRNA-rpl2-3 OTHER 0.001227 0.998773
- Relevant literature
Remove functions
Remove functional annotation features from the graph database. Functional annotations include the GO, pfam, tigrfam and interpro nodes as well as mRNA node properties for COG, phobius and signalp. There are multiple modes available:
‘all’ removes all functional annotation nodes and properties.
‘nodes’ removes all GO, pfam, tigrfam and interpro nodes.
‘properties’ removes all COG, phobius and signalp properties from mRNA nodes.
‘COG’ removes all COG properties from mRNA nodes.
‘phobius’ removes all phobius properties from mRNA nodes.
‘signalp’ removes all signalp properties from mRNA nodes.
‘bgc’ removes all AntiSMASH BGC nodes and relationships.
- Parameters
<databaseDirectory>
Path to the database root directory.
- Options
--mode/-mMode for which annotations to remove (default: all). Can be one of ‘all’, ‘nodes’, ‘properties’, ‘COG’, ‘phobius’ or ‘signalp’, ‘bgc’. See above for more information.
- Example commands
$ pantools remove_functions tomato_DB $ pantools remove_functions --mode nodes tomato_DB
Add antiSMASH
Read antiSMASH output and incorporate Biosynthetic Gene Clusters (BGC) nodes into the pangenome database. A ‘bgc’ node holds the gene cluster product, the cluster address and has a relationship to all gene nodes of the cluster. For this function to work, antiSMASH should be performed with the same FASTA and GFF3 files used for building the pangenome. antiSMASH output will not match the identifiers of the pangenome when no GFF file was included.
As of PanTools v3.3.4 the required antiSMASH version is 6.0.0. Gene cluster information is parsed from the .JSON file that is generated in each run. We try to keep the parser updated with newer versions but please contact us when this is no longer the case.
Version |
Version Date |
|
|---|---|---|
antiSMASH |
6.0.0 |
21-02-2021 |
- Parameters
<databaseDirectory>
Path to the database root directory.
<antiSMASHFile>
A text file with on each line a genome number and the full path to the corresponding antiSMASH output file, separated by a space.
- Options
--annotations-file/-AA text file with the identifiers of annotations to be included, each on a separate line. The most recent annotation is selected for genomes without an identifier.
- Example antiSMASH file
The <antiSMASHFile> requires to be formatted like a regular annotation input file. Each line of the file starts with the genome number followed by the full path to the JSON file.
1 /mnt/scratch/IPO3844/antismash/IPO3844.json 4 /home/user/IPO3845/antismash/IPO3845.json
- Example commands
$ pantools add_antismash tomato_DB clusters.txt $ pantools add_antismash -A annotations.txt tomato_DB clusters.txt
Function overview
Creates several summary files for each type of functional annotation present in the database: GO, PFAM, InterPro, TIGRFAM, COG, Phobius, and biosynthetic gene clusters from antiSMASH. In addition to the functions that must be added via add_functions, this function also requires proteins to be clustered by group.
- Parameters
<databaseDirectory>
Path to the pangenome database root directory.
- Options
--include/-iOnly include a selection of genomes.
--exclude/-eExclude a selection of genomes.
--annotations-file/-AA text file with the identifiers of annotations that should be used. The most recent annotation is selected for genomes without an identifier.
- Example commands
$ pantools function_overview tomato_DB $ pantools function_overview --include=2-4 tomato_DB
- Output
Output files are written to function directory in the database. The overview CSV files are tables with on each row a function identifier with the frequency of per genome and.
functions_per_group_and_mrna.csv, overview of all homology groups and the associated functions.
function_counts_per_group.csv,
go_overview.csv, overview of the GO terms in the pangenome.
pfam_overview.csv, overview of the PFAM domains in the pangenome.
tigrfam_overview.csv, overview of the TIGRFAMs in the pangenome.
interpro_overview.csv, overview of the InterPro domains in the pangenome.
bgc_overview.csv, overview of the added biosynthetic gene clusters from antiSMASH in the pangenome.
phobius_signalp_overview.csv, overview of the included Phobius transmembrane topology and signal peptide predictions in the pangenome.
cog_overview.csv, overview of the functional COG categories in the pangenome.
cog_per_class.R, an R script to plot the distribution of COG categories over the core, accessory, unique homology groups.
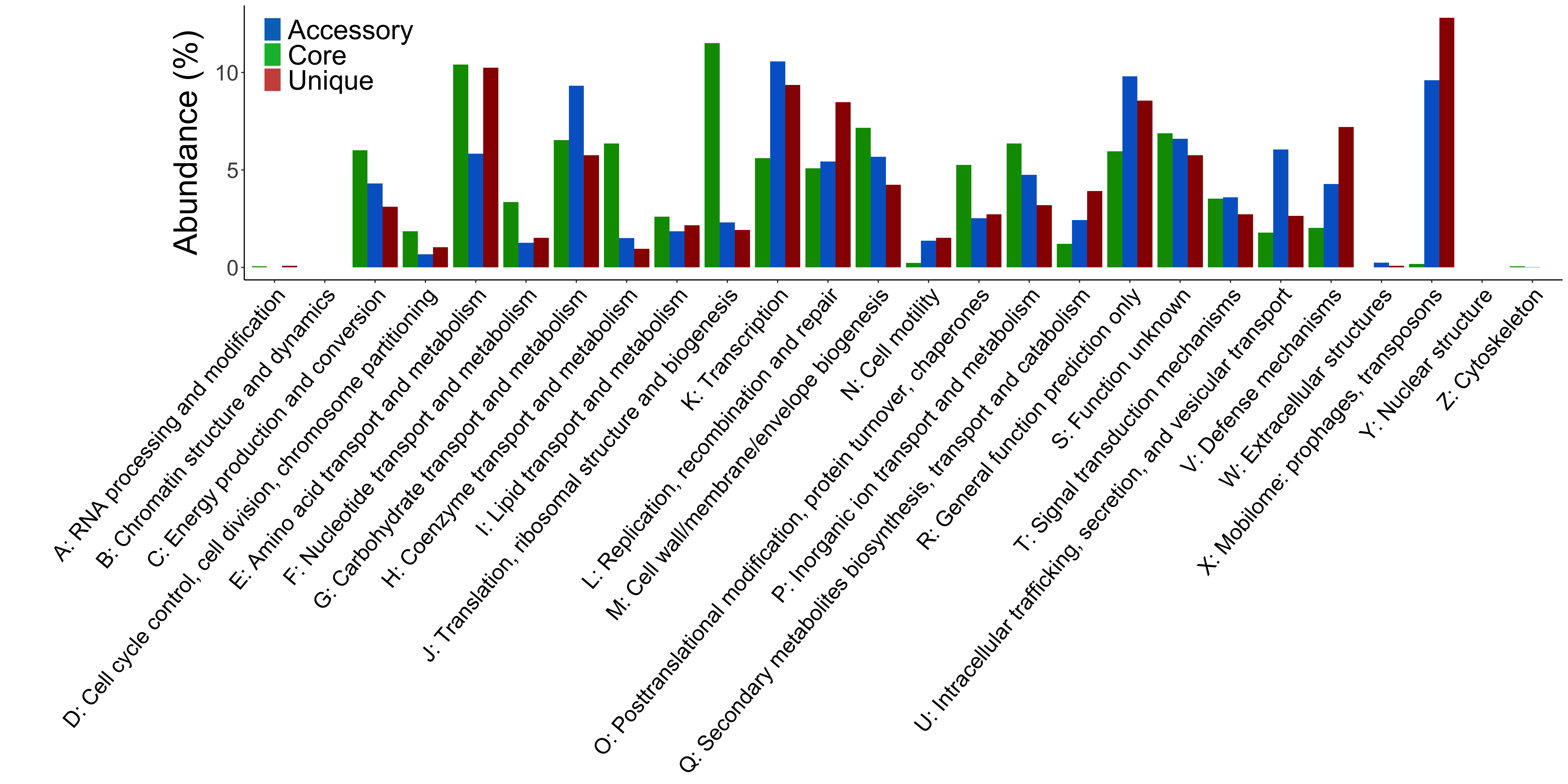
Fig. 10 Example output of cog_per_class.R. The proportion of COGs functional categories assigned to homology groups.
Phenotypes
Add phenotypes
Including phenotype data to the pangenome which allows the identification of phenotype specific genes, SNPs, functions, etc.. Altering the data is done by rerunning the command with an updated CSV file.
Values recognized as round number are converted to an Integer and to a Double when having one or multiple decimals.
Boolean types are identified by checking if the value matches ‘true’ or ‘false’, ignoring capitalization of letters.
String values remain completely unaltered except for spaces and quotes characters. Spaces are changed into an underscore (’_’) character and quotes are completely removed.
--bins. For skewed data,
consider making the bins manually and include this as string
phenotype.- Parameters
<databaseDirectory>
Path to the database root directory.
<phenotypesFile>
A CSV file containing the phenotype information.
- Options
--scratch-directoryTemporary directory for storing localization update files. If not set a temporary directory will be created inside the default temporary-file directory. On most Linux distributions this default temporary-file directory will be
/tmp, on MacOS typically/var/folders/.If a scratch directory is set, it will be created if it does not exist. If it does exist, PanTools will verify the directory is empty and, if not, raise an exception.
--appendDo not remove existing phenotype nodes but only add new properties to them. If a property already exists, values from the new file will overwrite the old.
--binsNumber of bins used to group numerical values of a phenotype (default: 3).
- Example phenotypes file
The input file needs to be in .CSV format, a plain text file where each value is separated by a comma. The first row should start with ‘Genome,’ followed by the phenotype names and/or identifiers. The first column must start with genome numbers corresponding to the one in your pangenome. Phenotypes and metadata must be placed on the same line as their genome number. A field can remain empty when the phenotype for a genome is missing or unknown. Here below is an example of five genomes contains six phenotypes:
Genome,Gram,Region,Pathogenicity,Boolean,float,species 1,+,NL,3,True,0.1,Species 2,+,BE,,False,0.1,Species3 3,+,LUX,7,true,0.1,Species3 4,+,NL,9,false,0.1,Species3 5,+,BE,15,TRUE,0.1,Species1
- Example commands
$ pantools add_phenotypes tomato_DB pheno.csv $ pantools add_phenotypes --append tomato_DB pheno.csv
- Output
Phenotype information is stored in ‘phenotype’ nodes in the graph. An output file is written to the database directory.
phenotype_overview.txt, a summary of the available phenotypes in the pangenome.
Remove phenotypes
Delete phenotype nodes or remove specific phenotype information from
the nodes. The specific phenotype property needs to be specified with
--phenotype. When this argument is not included, phenotype nodes
are removed.
- Parameters
<databaseDirectory>
Path to the database root directory.
- Options
--include/-iOnly remove nodes of the selected genomes.
--exclude/-eDo not remove nodes of the selected genomes.
--phenotype/-pName of the phenotype. All information of the given phenotype is removed from ‘phenotype’ nodes.
- Example commands
$ pantools remove_phenotypes tomato_DB $ pantools remove_phenotypes --phenotype=color tomato_DB $ pantools remove_phenotypes --phenotype=color --exclude=11,12 tomato_DB
Genomic variation
Add genomic variation to the pangenome database. These functions can handle SNP (single nucleotide polymorphism)/InDel (insertion/deletion) and PAV (presence/absence variation) information but will only consider genic variation when adding the information to the database. For SNP/InDel information, VCF (variant call format) files are required. For PAV information, a tab-separated file with 1s and 0s describing the presence and absence, respectively.
Add Variants
Add variants to the pangenome database. The function will only consider genomic variation that is present in the mRNA features of the pangenome. The SNP/InDel information will be used to create a consensus sequence for each mRNA features. For each accession and mRNA features, a new variant node will be created to hold this consensus sequence.
Several temporary files will be created during the process: a fasta file
containing the original mRNA sequences and fasta files containing the consensus
mRNA sequences for each sample. These files will be deleted after the process
is finished unless the --keep-intermediate-files option is used.
By default, the location of these files will be at /tmp for Linux and
/var/folders for macOS. The location can be changed with the
--scratch-directory option.
NB: VCF files that are not indexed with tabix will be indexed automatically on their original location!
- Required software
- Parameters
<databaseDirectory>
Path to the database root directory.
<vcfsFile>
A text file with on each line a genome number and the full path to a corresponding VCF file, separated by a space.
- Options
--threads/-tNumber of threads to use. Default: total number of cores available or 8, whichever is lower.
--scratch-directoryTemporary directory for storing intermediate files. If not set a temporary directory will be created inside the default temporary-file directory. On most Linux distributions this default temporary-file directory will be
/tmp/, on MacOS typically/var/folders/.If a scratch directory is set, it will be created if it does not exist. If it does exist, PanTools will verify the directory is empty and, if not, raise an exception.
--keep-intermediate-filesKeep intermediate consensus fasta and corresponding log files.
- Example VCFs file list
1 /path/to/LA1547.vcf.gz 1 /path/to/LA1557.vcf.gz 4 /path/to/LA1582.vcf.gz
- Example commands
$ pantools add_variants tomato_DB vcf_locations.txt $ pantools add_variants -t 4 tomato_DB vcf_locations.txt
Remove variants
Remove variants from the pangenome database. This function will remove all
VCF information from the database. All variant nodes created by the
add_variants function will be removed. The VCF information will be
removed from the accession nodes. If there is no variant information
left for an accession node, the node will be removed.
- Parameters
<databaseDirectory>
Path to the database root directory.
- Example commands
$ pantools remove_variants tomato_DB
Add PAVs
Add PAVs to the pangenome database. PAV information can only be added about mRNA features. For each accession and mRNA feature, PAV information can be stores in the database. Only values of 1 and 0 are allowed in the PAV file. A value of 1 indicates that the gene is present in the sample and a value of 0 indicates that the gene is absent in the sample.
- Parameters
<databaseDirectory>
Path to the database root directory.
<pavsFile>
A text file with on each line a genome number and the full path to a corresponding PAV file, separated by a space.
- Example PAVs file list
1 /path/to/LA1547.pav.tsv 4 /path/to/LA1582.pav.tsv
- Example PAV file
mrnaID accession102 accession103 accession104 LA1547_00001 1 1 1 LA1547_00002 1 1 0 LA1547_00003 1 1 1 LA1547_00004 1 0 1 LA1547_00005 1 1 1 LA1547_00006 0 0 1 LA1547_00007 0 0 0
- Example commands
$ pantools add_pavs tomato_DB pav_locations.txt
Remove PAVs
Remove PAVs from the pangenome database. This function will remove all
PAV information from the database. All variant nodes created by the
add_pavs function will be removed. The PAV information will be
removed from the accession nodes. If there is no variant information
left for an accession node, the node will be removed.
- Parameters
<databaseDirectory>
Path to the database root directory.
- Example commands
$ pantools remove_pavs tomato_DB
Variation overview
Create a readable overview of the variation in the pangenome database. The overview will be written to a text file. Per genome, this overview will contain the number of genes with PAV and/or VCF information and their sample names.
- Parameters
<databaseDirectory>
Path to the database root directory.
- Example commands
$ pantools variation_overview tomato_DB
- Output
The output file will be written to the variation directory in the database as a text file.
variation_overview.txt, a summary of available variation in the pangenome.
Phased pangenomics
Add phasing
Warning
This is a novel function and has not yet undergone testing by external users. Please report any bugs or issues to the PanTools team so we can improve it.
Include phasing information into the pangenome. A chromosome number combined with a phasing letter makes a phasing identifier. (Currently) a phasing identifier must be unique, therefore phasing related PanTools functionalities may only be useful when using chromosome scale and fully phased assemblies.
- Parameters
<databaseDirectory>
Path to the database root directory.
<phasingFile>
A text file with phasing information of sequences.
- Options
--assume-unphasedAll chromosomes without a letter will be be considered unphased.
- Example commands
$ pantools add_phasing tomato_DB phasing_info.txt
- Example input
The text file should have two columns, separated by a tab, space or comma. The first column can only contain sequence identifiers. The second column can be formatted in two different ways.
Input format 1. Chromosome numbersThe second colum contains only (chromosome) numbers. This number becomes the chromosome number. To obtain the phasing letters, we count the number sequences from the same genome within one cluster. The sequence order determines the phasing letter.Taking the example below, for the second chromosome: genome 1 has 4 sequences, genome 2 has 3 sequences, and genome 3 has 1 sequence. The assigned identifiers are:
Genome 1 - 2_A, 2_B, 2_C, 2_D
Genome 2 - 2_A, 2_B, 2_C
Genome 3 - 2_unphased
1_1 1 1_2 1 1_3 1 1_4 1 2_1 1 2_2 1 2_3 1 2_4 1 3_1 1 1_5 2 1_6 2 1_7 2 1_8 2 2_5 2 2_6 2 2_7 2 3_2 2
This file format is generated by running TreeCluster.py on a sequence-level k-mer distance tree.
$ TreeCluster.py -i sequence_kmer_distance.tree -m avg_clade -t 0.03 > phasing_info.txt
Input format 2. Directly assign identifiersExample file that will directly assign phasing identifiers to sequences. The identifiers are identical to the example above.1_1,1_A 1_2,1_B 1_3,1_C 1_4,1_D 2_1,1_A 2_2,1_B 2_3,1_C 2_4,1_D 3_1,unphased 1_5,2_A 1_6,2_B 1_7,2_C 1_8,2_D 2_5,2_A 2_6,2_B 2_7,2_C 3_2,unphased
Repetitive elements
Add repeats
Warning
This is a novel function and has not yet undergone testing by external users. Please report any bugs or issues to the PanTools team so we can improve it.
Add repeat annotations to an existing pangenome. PanTools is only able to read General Feature Format (GFF) files. Reads everything as a single line thus ignores hierarchical levels of the GFF format. Repeat ‘type’ is based on the 3rd column.
- Parameters
<databaseDirectory>
Path to the database root directory.
<annotationsFile>
A text file with on each line a genome number and the full path to the corresponding annotation file, separated by a space.
- Options
--connectConnect the annotated genomic features to nucleotide nodes in the DBG.
--strictStop the annotation if sequences or repeat coordinates do not match to the database.
- Example commands
$ pantools add_repeats tomato_DB repeats.txt $ pantools add_repeats potato_DB repeats.txt --connect --strict
- Example input file
In the required input file each line starts with the genome number followed by the full path to a GFF file, separated by a space.
1 /always/genome1.gff 2 /use_the/genome2.gff3 3 /full_path/genome3.gff
The GFF format consists of one line per feature, each containing 9 columns of data (plus optional track definition lines), that must be tab separated. Currently, we identify the repeat type through the 3rd column.
##seqid source sequence_ontology start end score strand phase attributes chr1A EDTA repeat_region 350 8207 . ? . ID=repeat_region_1;Name=TE_00012440;Classification=LTR/unknown;Sequence_ontology=SO:0000657;ltr_identity=0.9995;Method=structural;motif=TGCA;tsd=CCTGG chr1A EDTA target_site_duplication 350 354 . ? . ID=lTSD_1;Parent=repeat_region_1;Name=TE_00012440;Classification=LTR/unknown;Sequence_ontology=SO:0000434;ltr_identity=0.9995;Method=structural;motif=TGCA;tsd=CCTGG chr1A EDTA long_terminal_repeat 355 2216 . ? . ID=lLTR_1;Parent=repeat_region_1;Name=TE_00012440;Classification=LTR/unknown;Sequence_ontology=SO:0000286;ltr_identity=0.9995;Method=structural;motif=TGCA;tsd=CCTGG chr1A EDTA LTR_retrotransposon 355 8202 . ? . ID=LTRRT_1;Parent=repeat_region_1;Name=TE_00012440;Classification=LTR/unknown;Sequence_ontology=SO:0000186;ltr_identity=0.9995;Method=structural;motif=TGCA;tsd=CCTGG chr1A EDTA helitron 2843 3627 4150 + . ID=TE_homo_0;Name=TE_00001861;Classification=DNA/Helitron;Sequence_ontology=SO:0000544;Identity=0.819;Method=homology chr1A EDTA helitron 3812 3914 360 + . ID=TE_homo_1;Name=TE_00001914;Classification=DNA/Helitron;Sequence_ontology=SO:0000544;Identity=0.822;Method=homology chr1A EDTA Mutator_TIR_transposon 5076 5627 4956 + . ID=TE_homo_2;Name=TE_00010497;Classification=DNA/DTM;Sequence_ontology=SO:0002280;Identity=0.985;Method=homology chr1A EDTA hAT_TIR_transposon 5801 6148 3156 - . ID=TE_homo_3;Name=TE_00003074;Classification=DNA/DTA;Sequence_ontology=SO:0002279;Identity=0.997;Method=homology chr1A EDTA long_terminal_repeat 6342 8202 . ? . ID=rLTR_1;Parent=repeat_region_1;Name=TE_00012440;Classification=LTR/unknown;Sequence_ontology=SO:0000286;ltr_identity=0.9995;Method=structural;motif=TGCA;tsd=CCTGG chr1A EDTA target_site_duplication 8203 8207 . ? . ID=rTSD_1;Parent=repeat_region_1;Name=TE_00012440;Classification=LTR/unknown;Sequence_ontology=SO:0000434;ltr_identity=0.9995;Method=structural;motif=TGCA;tsd=CCTGG chr1A EDTA Gypsy_LTR_retrotransposon 8203 8764 5107 + . ID=TE_homo_4;Name=TE_00012288_INT;Classification=LTR/Gypsy;Sequence_ontology=SO:0002265;Identity=0.993;Method=homology chr1A EDTA LTR_retrotransposon 8865 10542 11862 - . ID=TE_homo_5;Name=TE_00009031_LTR;Classification=LTR/unknown;Sequence_ontology=SO:0000186;Identity=0.932;Method=homology chr1A EDTA Copia_LTR_retrotransposon 10643 10979 2849 + . ID=TE_homo_6;Name=TE_00005676_LTR;Classification=LTR/Copia;Sequence_ontology=SO:0002264;Identity=0.967;Method=homology chr1A EDTA CACTA_TIR_transposon 10978 11061 501 + . ID=TE_homo_7;Name=TE_00006381;Classification=DNA/DTC;Sequence_ontology=SO:0002285;Identity=0.866;Method=homology
Repeat overview
Calculate the frequency and overlap of repeats in the genome (split into windows) and gene regions.
- Parameters
<databaseDirectory>
Path to the database root directory.
- Options
--include/-iOnly include a selection of genomes. This automatically lowers the threshold for core genes.
--exclude/-eExclude a selection of genomes. This automatically lowers the threshold for core genes.
-—selection-fileText file with rules to use a specific set of genomes and sequences. This automatically lowers the threshold for core genes.
--window-lengthSet the window length (default: 50000).
--upstreamSet the gene upstream region (default: 1000).
--downstreamSet the gene downstream region (default: 1000).
--exclude-repeatsText file to only include (or exclude) certain repeat types for the analysis.
- Example commands
$ pantools repeat_overview tomato_DB $ pantools repeat_overview tomato_DB --selection-file sequence_selection.txt $ pantools repeat_overview tomato_DB --window-length 1000000 --upstream 5000 --downstream 5000
- Example input files
The
--selection-filemust be a single line text file to include or exclude a selection of repeat types. The repeat types must be separated through commas.INCLUDE = LTR_retrotransposon, LINE_element, Copia_LTR_retrotransposon
EXCLUDE = Gypsy_LTR_retrotransposon
- Output
Output files are written to the repeats directory in the database.
windows_all_sequences.csv, Holds the calculated repeat frequency and bases overlapped per repeat type for all sequences combined.
statistics_genomes_sequences.csv, per genome and sequence, holds the calculated repeat frequency and bases overlapped per repeat type and all repeat types combined.
repeats_in_genes.csv provides repeat statistics for individual genes.
coverage_plot.R creates a coverage plot for each sequence.
coverage_plot.R creates a coverage plot for every sequence pair.
density_plot.R creates a density and density abundance plot for each sequence.
density_plot_two_sequences.R creates a density and % density plot for every sequence pair.
Additional output files named after each sequence identifier are available in the repeats/windows directory. Per window, these hold the calculated repeat frequency and bases overlapped per repeat type and all repeat types combined.
Removing data
Remove nodes
Remove a selection of nodes and their relationships from the pangenome. For a pangenome database the following nodes should never be removed: nucleotide, pangenome, genome, sequence. When using a panproteome, mRNA nodes cannot be removed.
- Parameters
<databaseDirectory>
Path to the database root directory.
- Options
Requires one of
--nodes|--label,includeandexcludeonly work for--label.--include/-iOnly remove nodes of the selected genomes.
--exclude/-eDo not remove nodes of the selected genomes.
--nodes/-nOne or multiple node identifiers, separated by a comma.
--labelA node label, all nodes matching the label are removed.
- Example commands
$ pantools remove_nodes --nodes=10348734,10348735,10348736 tomato_DB $ pantools remove_nodes --label=busco --include=2-6 tomato_DB